m2ST: dual multi-scale graph clustering for spatially resolved transcriptomics
Published in Bioinformatics 2025, 2025
Wei Zhang, Ziqi Zhang, Hailong Yang, Te Zhang, Shu Jiang, Ning Qiao, Zhaohong Deng, Xiaoyong Pan, Hong-Bin Shen, Dong-Jun Yu, Shitong Wang. Bioinformatics 2025 (CCF B). https://doi.org/10.1093/bioinformatics/btaf221
m2ST
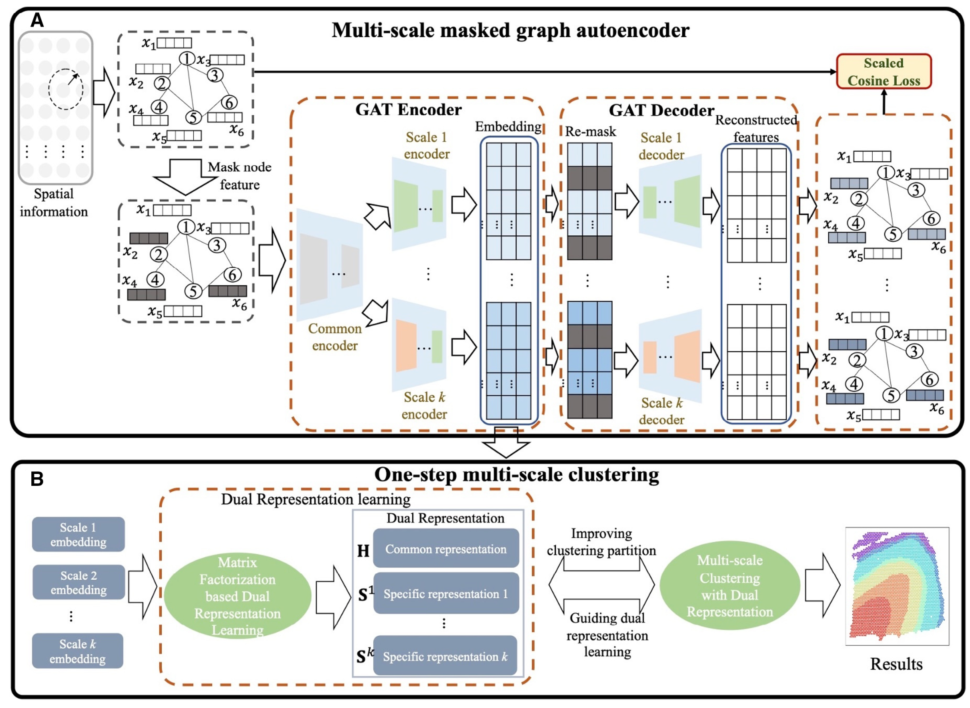
Motivation: Spatial clustering is a key analytical technique for exploring spatial transcriptomics data. Recent graph neural network-based methods have shown promise in spatial clustering but face notable challenges. One significant issue is that analyzing the functions and complex mechanisms of organisms from a single scale is difficult and most methods focus exclusively on the single-scale representation of transcriptomic data, potentially limiting the discriminative power of extracted features for spatial domain clustering. Furthermore, classical clustering algorithms are often applied directly to latent representation, making it a worthwhile endeavor to explore a tailored clustering method to further improve the accuracy of spatial domain annotation.
Results: To address these limitations, we propose m2ST, a novel dual multi-scale graph clustering method. m2ST first uses a multi-scale masked graph autoencoder to extract representations across different scales from spatial transcriptomic data. To effectively compress and distill meaningful knowledge embedded in the data, m2ST introduces a random masking mechanism for node features and uses a scaled cosine error as the loss function. Additionally, we introduce a tailored multi-scale clustering framework that integrates scale-common and scalespecific information exploration into the clustering process, achieving more robust annotation performance. Shannon entropy is finally utilized to dynamically adjust the importance of different scales. Extensive experiments on multiple spatial transcriptomic datasets demonstrate the superior performance of m2ST compared to existing methods